The thing genetic algorithms do efficiently—Parallel Effect Search —is not difficult to describe or demonstrate. It’s been under our noses since schemata and schema partitions were defined as concepts, but for various reasons, which I won’t get into here, it’s not widely appreciated. This blog post provides a quick introduction. For a more in depth treatment check out my FOGA 2013 paper Explaining Optimization in Genetic Algorithms with Uniform Crossover (slides here).
Schema Partitions and Effects
Let’s begin with a quick primer on schemata and schema partitions. Let 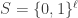 be a search space consisting of binary strings of length
be a search space consisting of binary strings of length  . Let
. Let 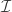 be some set of indices between
be some set of indices between 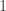 and
and  , i.e.
, i.e. 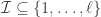 . Then
. Then 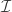 represents a partition of
represents a partition of  into
into 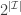 subsets called schemata (singular schema) as in the following example: suppose
subsets called schemata (singular schema) as in the following example: suppose 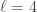 , and
, and  , then
, then 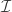 partitions
partitions  into four schemata:
into four schemata:



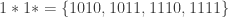
where the symbol  stands for ‘wildcard’. Partitions of this type are called schema partitions. As we’ve already seen, schemata can be expressed using templates, for example,
stands for ‘wildcard’. Partitions of this type are called schema partitions. As we’ve already seen, schemata can be expressed using templates, for example,  . The same goes for schema partitions. For example
. The same goes for schema partitions. For example  denotes the schema partition represented by the index set
denotes the schema partition represented by the index set 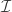 . Here the symbol
. Here the symbol 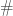 stands for ‘defined bit’. The order of a schema partition is simply the cardinality of the index set that defines the partition (in our running example, it is
stands for ‘defined bit’. The order of a schema partition is simply the cardinality of the index set that defines the partition (in our running example, it is  ). Clearly, schema partitions of lower order are coarser than schema partitions of higher order.
). Clearly, schema partitions of lower order are coarser than schema partitions of higher order.
Let us define the effect of a schema partition to be the variance of the average fitness values of the constituent schemata under sampling from the uniform distribution over each schema. So for example, the effect of the schema partition  is
is
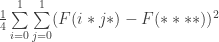
where the operator  gives the average fitness of a schema under sampling from the uniform distribution.
gives the average fitness of a schema under sampling from the uniform distribution.
You’re now well poised to understand Parallel Effect Search. Before we get to a description, a brief detour to provide some motivation: We’re going to do a thought experiment in which we examine how effects change with the coarseness of schema partitions. Let ![[\mathcal I]](https://s0.wp.com/latex.php?latex=%5B%5Cmathcal+I%5D&bg=ffffff&fg=333333&s=0&c=20201002) denote the schema partition represented by some index set
denote the schema partition represented by some index set 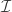 . Consider a search space
. Consider a search space 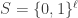 with
with  , and let
, and let  . Then
. Then ![[\mathcal I]](https://s0.wp.com/latex.php?latex=%5B%5Cmathcal+I%5D&bg=ffffff&fg=333333&s=0&c=20201002) is the finest possible partition of
is the finest possible partition of  ; one where each schema in the partition has just one point. Consider what happens to the effect of
; one where each schema in the partition has just one point. Consider what happens to the effect of ![[\mathcal I]](https://s0.wp.com/latex.php?latex=%5B%5Cmathcal+I%5D&bg=ffffff&fg=333333&s=0&c=20201002) as we start removing elements from
as we start removing elements from 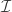 . It should be relatively easy to see that the effect of
. It should be relatively easy to see that the effect of ![[\mathcal I]](https://s0.wp.com/latex.php?latex=%5B%5Cmathcal+I%5D&bg=ffffff&fg=333333&s=0&c=20201002) decreases monotonically. Why? Because we’re averaging over points that used to be in separate partitions. Don’t proceed further until you convince yourself that coarsening a partition tends to decrease its effect.
decreases monotonically. Why? Because we’re averaging over points that used to be in separate partitions. Don’t proceed further until you convince yourself that coarsening a partition tends to decrease its effect.
Finally, observe that the number of schema partitions of order  is
is  . So for
. So for  , the number of schema partitions of order 2,3,4 and 5 are on the order of
, the number of schema partitions of order 2,3,4 and 5 are on the order of  , and
, and  respectively. The take away from our thought experiment is this: while a search space may have vast numbers of coarse schema partitions, most of them will have negligible effects (due to averaging). In other words, while coarse schema partitions are numerous, ones with non-negligible effects are rare.
respectively. The take away from our thought experiment is this: while a search space may have vast numbers of coarse schema partitions, most of them will have negligible effects (due to averaging). In other words, while coarse schema partitions are numerous, ones with non-negligible effects are rare.
So what exactly does a genetic algorithm do efficiently? Using experiments and symmetry arguments I’ve demonstrated that a genetic algorithm with uniform crossover can sift through vast numbers of coarse schema partitions in parallel and identify partitions with non-negligible effects. In other words, a genetic algorithm with uniform crossover can implicitly perform multitudes of effect/no-effect multifactor analyses and can efficiently identify interacting loci with non-negligible effects.
Let’s Play a Game
It’s actually quite easy to visualize a genetic algorithm as it identifies such loci. Let’s play a game. Consider a stochastic function that takes bitstrings of length 200 as input and returns an output that depends on the values of the bits of at just four indices. These four indices are fixed; they can be any one of the  combinations of four indices between between 1 and 200. Given some bitstring, if the parity of the bits at these indices is 1 (i.e. if the sum of the four bits is odd) then the stochastic function returns a value drawn from the magenta distribution (see below). Otherwise, it returns a value drawn from the black distribution. The four indices are said to be pivotal. All other indices are said to be non-pivotal.
combinations of four indices between between 1 and 200. Given some bitstring, if the parity of the bits at these indices is 1 (i.e. if the sum of the four bits is odd) then the stochastic function returns a value drawn from the magenta distribution (see below). Otherwise, it returns a value drawn from the black distribution. The four indices are said to be pivotal. All other indices are said to be non-pivotal.

As per the discussion in the first part of this post, the set of pivotal indices is the dual of a schema partition of order 4. Of all the schema partitions of order 4 or less, only this partition has a non-zero effect. All other schema partitions of order 4 or less have no effect. (Verify for yourself that this is true) In other words parity seems like a Needle in a Haystack (NIAH) problem—a problem, in other words, that requires brute force.
Now for the rules of the game: Say I give you query access to the stochastic function just described, but I do not tell you what four indices are pivotal. You are free to query the function with any bitstring 200 bits long as many times as you want. Your job is to recover the pivotal indices I picked, i.e. to identify the only schema partition of order 4 or less with a non-negligible effect.
Take a moment to think about how you would do it? What is the time and query complexity of your method?
What “Not Breaking a Sweat” Looks Like
The animation below shows what happens when a genetic algorithm with uniform crossover is applied to the stochastic function just described. Each dot displays the proportion of 1’s in the population at a locus. Note that it’s trivial to just “read off” the proportion of 0s at each locus. The four pivotal loci are marked by red dots. Of course, the genetic algorithm does not “know” that these loci are special. It only has query access to the stochastic function.
As the animation shows, after 500 generations you can simply “read off” the four loci I picked by examining the proportion of 1s to 0s in the population at each locus. You’ve just seen Parallel Effect Search in action. The chromosome size in this case is 200, so there are  possible combinations of four loci. From all of these possibilities, the genetic algorithm managed to identify the correct one within five hundred generations.
possible combinations of four loci. From all of these possibilities, the genetic algorithm managed to identify the correct one within five hundred generations.
Let’s put the genetic algorithm through it’s paces. I’m going to tack on an additional 800 non-pivotal loci while leaving the indices of the four pivotal loci unchanged. Check out what happens:
[Note: more dots in the animation below does not mean a bigger population or more queries. More dots just means more loci under consideration. The population size and total number of queries remain the same]
So despite a quintupling in the number of bits, entailing an increase in the number of coarse schema partitions of order 4 to  , the genetic algorithm solves the problem with no increase in the number of queries. Not bad. (Of course we’re talking about a single run of a stochastic process. And yes, it’s a representative run. See chapter 3 of my dissertation to get a sense for the shape of the underlying process)
, the genetic algorithm solves the problem with no increase in the number of queries. Not bad. (Of course we’re talking about a single run of a stochastic process. And yes, it’s a representative run. See chapter 3 of my dissertation to get a sense for the shape of the underlying process)
Let’s take it up another notch, and increase the length of the bitstrings to 10,000. So now we’re looking at  combinations of four loci. That’s on the order of a million trillion combinations. This time round, let’s also change the locations of the 4 pivotal loci. Will the genetic algorithm find them in 500 generations or less?
combinations of four loci. That’s on the order of a million trillion combinations. This time round, let’s also change the locations of the 4 pivotal loci. Will the genetic algorithm find them in 500 generations or less?
How’s that for not breaking a sweat? Don’t be deceived by the ease with which the genetic algorithm finds the answer. This is not an easy problem.
To run the experiments yourself download speedyGApy, and run it with
python speedyGA.py --fitnessFunction seap --bitstringLength <big number>
noting that the increase in “wall clock” time between generations as you increase bitstringLength is due to an increase in the running time of everything (including query evaluation). The number of queries (i.e. number of fitness evaluations), however, stays the same. To learn how a genetic algorithm parlays parallel effect search into a general-purpose global search heuristic called hyperclimbing, read my FOGA 2013 paper Explaining Optimization in Genetic Algorithms with Uniform Crossover (slides here).




